Help
(1) Submit a mRNA to get siRNAs with thier efficacy: Sequence should be in two line fasta format in which first line having ">name" and second line having sequence. Please see the example.
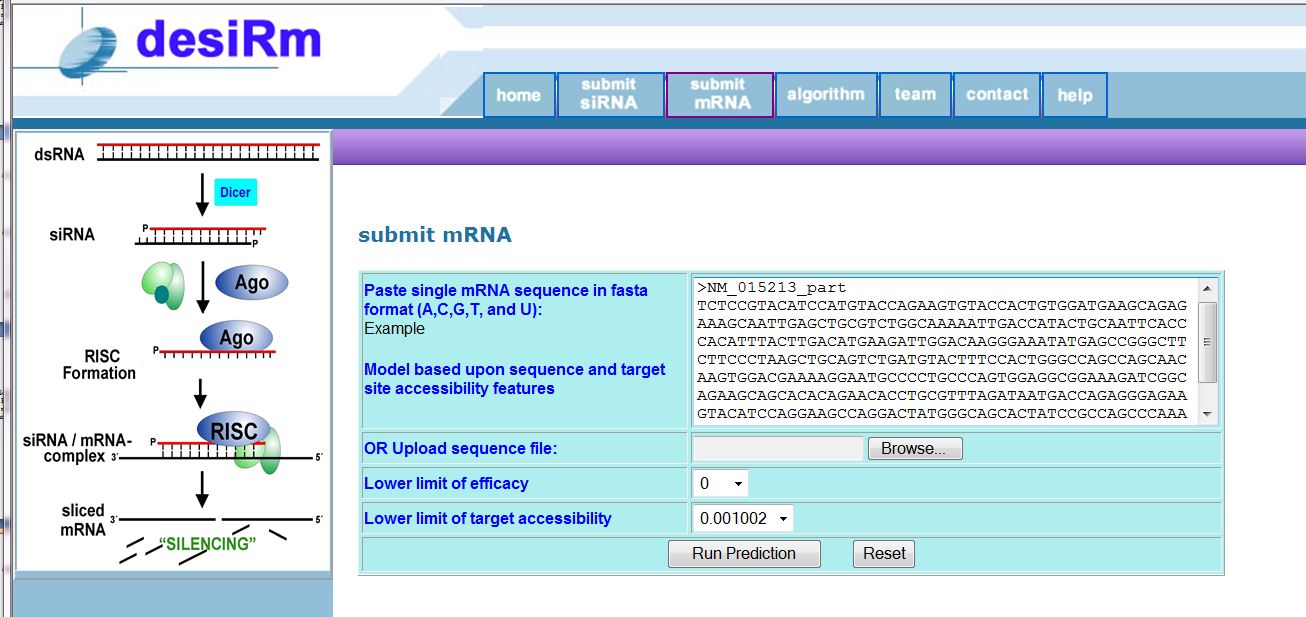
Figure 1: Submission fields of mRNA to get siRNAs.
Prediction results
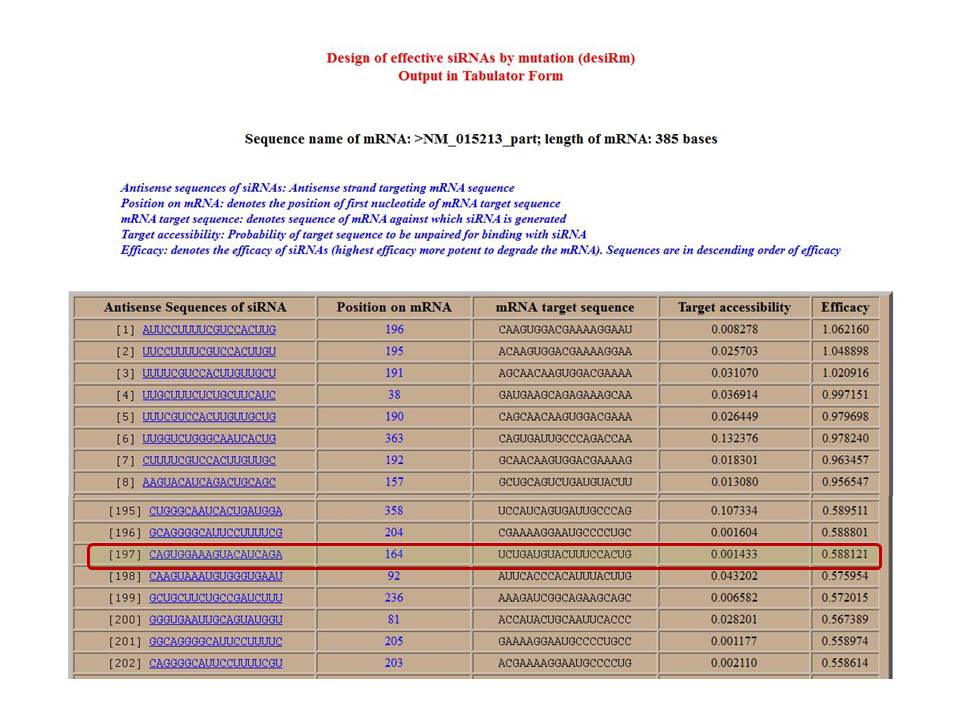
Figure 2: Snapshot of output result containing sequence of siRNA, target position, target sequence and accessibility with predicted efficacy. To improve the efficacy of 197th siRNA targeting on 164th position (highlighted), click this sequence.
To get single-mutated siRNAs
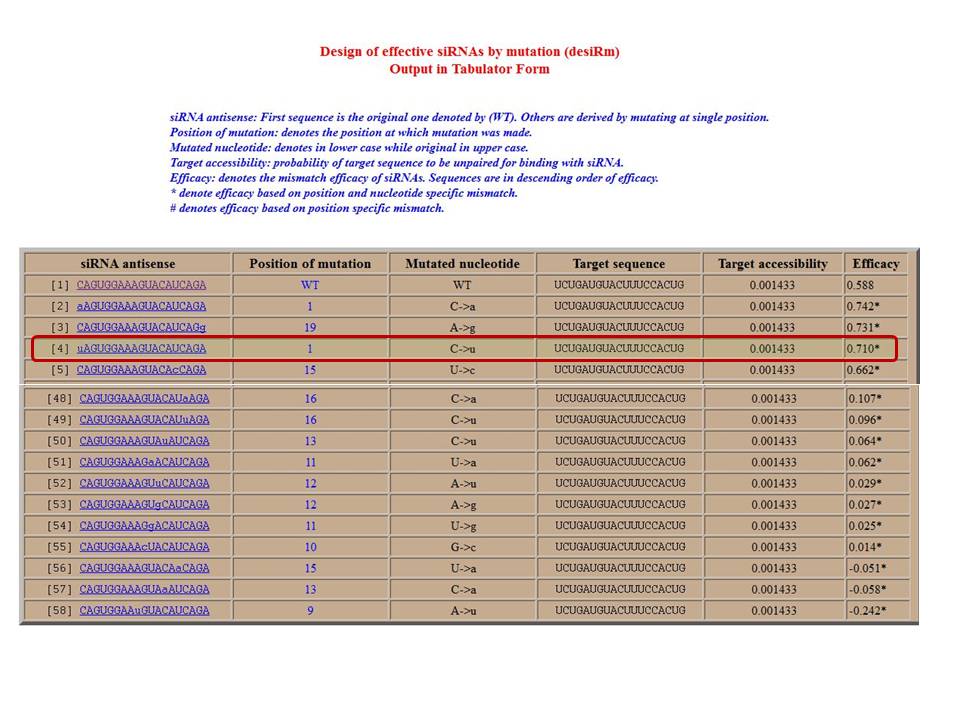
Figure 3: Snapshot of output result of single-mutated siRNA efficacy. Result contains mutated siRNA, position of mutation, type of mutation, target sequence and accessibility with predicted efficacy. First sequence (WT) is original, mutation at 1st nt in siRNA increase their efficacy to 0.710. For further improvement click the second siRNA.
To get double-mutated siRNAs, further click on siRNA
(2) Submit a siRNA to get mutated siRNAs with thier efficacy: Input should be 19 nt siRNA sequence with 19 nt target sequence separated by "::". All sequence should be written in 5 to 3 direction.
eg GGUCCACAUUCUAUUUUAA::UUAAAAUAGAAUGUGGACC
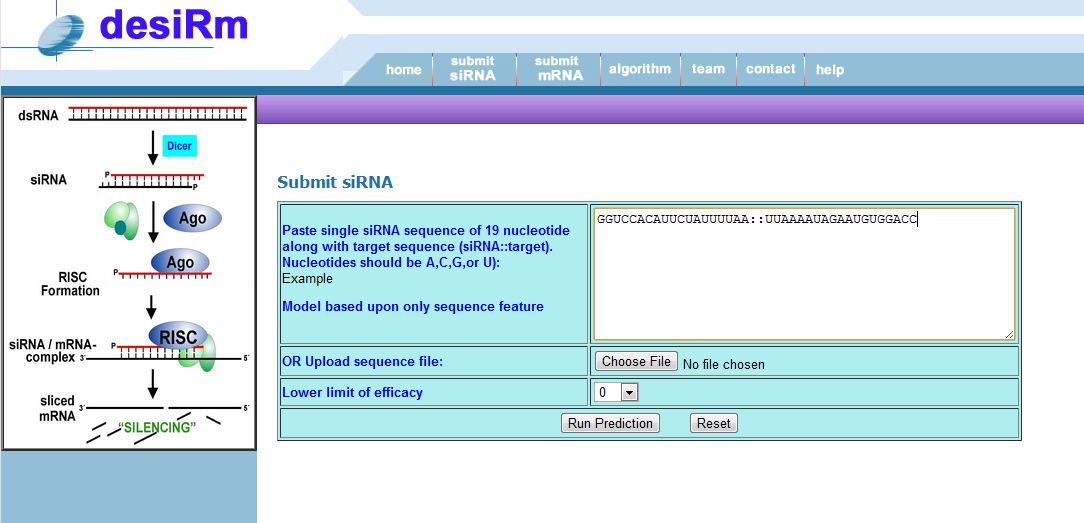
Figure 5: Submission fields of siRNA to get mutant siRNAs.
|
