|
Help for Pprint ::
Pprint is a Support Vectors Machine (SVM) based prediction method of RNA-interacting resiudes in a protein. The prediction is done by SVM model trained on PSSM profile generated by PSI-BLAST search of 'nr' protein database.
The submission form.
To submit a protein sequence for prediction, following steps have to be followed. The numebr in parenthesis indicates the corrosponding input box of submission form (displayed in Figure 1).
- Go to submit page of Pprint. click here to open submission page
- Enter name of sequence. (1) [optional]
- Put a valid e-mail id. (2) [optional]
At this id you receive an e-mail when job will be completed.
- Paste (3) /upload (4) FASTA format amino acid sequence.
- Select threshold of prediction. (5)
The probability of correct prediction directly depends on the threshold. For prediction with high confidence (less probability of false positive prediction) high threshold should be choosen. At low threshold, the probability of false negative prediction is very low. On default theshold (-0.2) the rate of false negative and false positive prediction was nearly equal during training of SVM model.
- In case you want to remove all enteries from submission page, click 'clear' tab. (6)
- Click 'Run Prediction' to submit protein for prediction.(7)
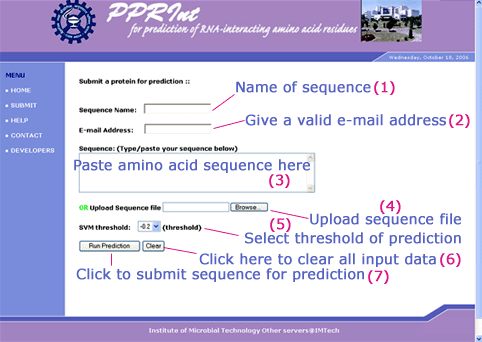
Figure 1: Different fields of Pprint web-server submission form.
The result.
The prediction result will be displayed in two section (Figure 2).
- Color coded amino acid sequence submitted for the prediction. The red coloured residues are predicted as RNA-interacting and blue coloured residues are predicted as non RNA-interacting.
- Below the amino acid sequence, residue-wise detail prediction is given in tabular format. This table contains three columns (i) amino acid residue, (ii) SVM score and (iii) prediction.
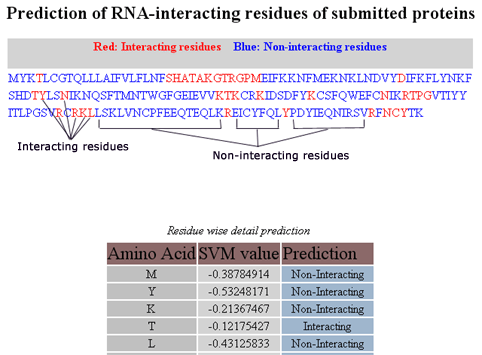
Figure 2: Prediction result of Pprint.
|

